- Research
- Publication
Published:
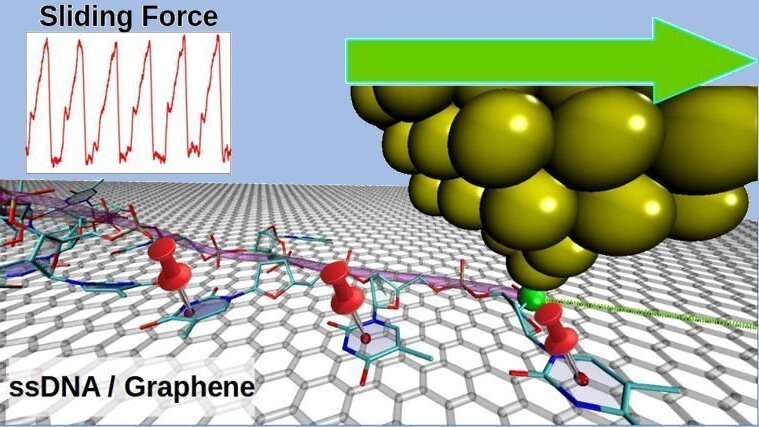
We have performed molecular dynamics simulations of nanomanipulation experiments on short single-stranded DNA chains elastically driven on a graphene surface. After a brief transient, reproducible stick-slip cycles are observed on chains made by 10 units of thymine, cytosine, adenine, and guanine. The cycles have the periodicity of the graphene substrate, and take place via an intermediate stage, appearing as a dip in the sawtooth variations of lateral force recorded while the chains are manipulated. Guanine presents remarkable differences from the other bases, since a lower number of nucleotides are prone to stick to the substrate in this case. Nevertheless, the magnitudes of static friction and lateral stiffness are similar for all chains (30 pN and 0.7 N/m per adsorbed nucleotide respectively).
Publication
J. G. Vilhena, Enrico Gnecco, Rémy Pawlak, Fernando Moreno-Herrero, Ernst Meyer, Rubén Pérez: "Stick-Slip Motion of ssDNA over Graphene", Journal of Physical Chemistry B 122 (2018) 840–846, DOI: 10.1021/acs.jpcb.7b06952External link